
A design matrix specifies a
statistical model consisting of a set of predictors
or explanatory variables.
The design matrix is, thus, an important tool to formulate hypotheses
about expected changes of the fMRI signal. Using the General Linear Model
(GLM), the statistical model specified in a design matrix is compared
with the measured time course at each voxel. The comparison of the model
and the data is expressed as an R or F value for each voxel which tells
how good the overall model fits
or explains the data. If the
R or F value of a voxel passes a statistical threshold, the respective
voxel will be highlighted by appropriate color-coding. In addition, the
GLM results in a set of beta weights
for each voxel telling which
predictors contribute significantly to explain a voxels time course.
For further details about the computations performed by the GLM, see section
"<GLM ??>".
In the following, it is described, how design matrices are constructed
for a single fMRI study (section "Single study design matrices")
as well as for arbitrary many studies in the context of multiple runs
of many subjects (section "Multi study design matrices"). As
an example, we will use data from the "ClockTask" folder. This
folder is located in the "Samples" directory of the BrainVoyager
CD; it might be also in your BrainVoyager folder of your hard disk (i.e.
"C:\Program Files\BrainVoyager\Samples\ClockTask") in case you
have selected this sample during program installation.
Single study design matrices
A design matrix for a single study (FMR or VTC) is typically defined interactively
by clicking on respective conditions of a stimulation protocol. Alternatively,
a respective RTC data file can be created outside of BrainVoyager. To
prepare the definition of a design matrix, open the "BS_TAL.vmr"
project file and link the "BS_run1_r1_pp.vtc" VTC file. To specify
a design matrix for the specified single run, click the General
Linear Model: Single Study... entry
in the Analysis menu. This will
invoke the Single Study General Linear Model
dialog as shown below:

The dialog shows a time line from left
to right representing the 126 measurement time points of the experiment.
The time line is segmented according to the stimulation protocol of the
study which the program found by following a reference to the protocol
saved within the VTC file. The study used as an example here is from an
event-related study of mental imagery, the "mental clock task"
(Trojano et al., 2000, Formisano et al., 2001). In an individual trial,
the subject hears.. In the protocol, each trial has been segmented into
3 periods, "Auditory stimulation" (red segments), "Clock
imagery" (green segments) and "Response" (blue segments).
A predictor can be graphically defined simply by clicking with the right
mouse button on a condition. After clicking with the right mouse button
on a red segment, the dialog will look as shown in the following figure:

Note that the Name: text field shows "Auditory stimulation" as the name of the predictor. This predictor name has been copied from the stimulation protocol and can be changed by editing the Name: field. In an event-related design with many short events, it might be difficult to hit the visual representation of the respective protocol segment. To increase the width of the segments, you can resize the dialog using the size grip at the right lower corner. Another alternative is to use the protocol itself. For this approach, invoke the Stimulation Protocol by clicking the Protocol button.

In the protocol window, you can click with the right mouse button on a condition name to set the respective segments in the Single Study General Linear Model dialog to "1" and with a click with the left mouse button to "0". The defined "box-car" function does not reflect the hemodynamic delay of the fMRI response. Click the HRF button to convolve the box-car properly with a hemodynamic response function:

Now we have defined a design matrix with one predictor. It is possible at any point to visualize the design matrix numerically. Click the Options button to invoke the Options dialog and then click the Design matrix button. This will present the following General Linear Model design matrix dialog:

In the design matrix, time runs from top
to bottom whereas in the Single Study
General Linear Model dialog, time runs from left to right. Each
column represents one predictor. The first column contains a numerical
representation of the graphically defined predictor with its name, "Auditory
stimulation", shown in the first row. The numerical values are the
results of the convolution of the box car function with the hemodynamic
response function. If you visualize the design matrix prior to pressing
the HRF button, you would see
only 1.0 and 0.0 values in the first column. The second predictor with
a constant value of "1" has been added automatically to the
design matrix and is necessary for the GLM to estimate the level of the
fMRI signal at a voxel. As we will see in the section about multi study
design matrices, such a constant predictor will be added automatically
for each study. You can use the scroll bar(s) to browse to any position
within the design matrix. You can also increase the size of the dialog
by using the size grip in the right lower corner.
A graphical representation similar to the one used in the SPM software
package can be produced by clicking the SPM
style option. In this representation, numerical values within a
cell are shown as grey levels: a value of 1.0 is colored white, a value
of -1.0 is colored black and intermediate values are colored with a respective
grey level. Since the "Mean" predictor has a constant value
of 1.0, all entries in the second column are colored white. The gradual
changes of predictor 1 (column 1) are shown with appropriate grey levels.
You can use the Zoom in and Zoom out buttons to enlarge or reduce
the size of the cells of the design matrix. In SPM style, you can click
the Zoom out button repeatedly
and still get a good representation of a complex design matrix.
Remark The design matrix is used exactly as shown in this dialog for a subsequent GLM computation. This holds also true for any complex multi subject/study design matrix. In order to know precisely what statistical model is constructed and used by BrainVoyager, you can always check the design matrix in this dialog. In addition, the actually used design matrix is also saved to disk automatically at the beginning of a GLM computation ("DesignMatrix.txt").
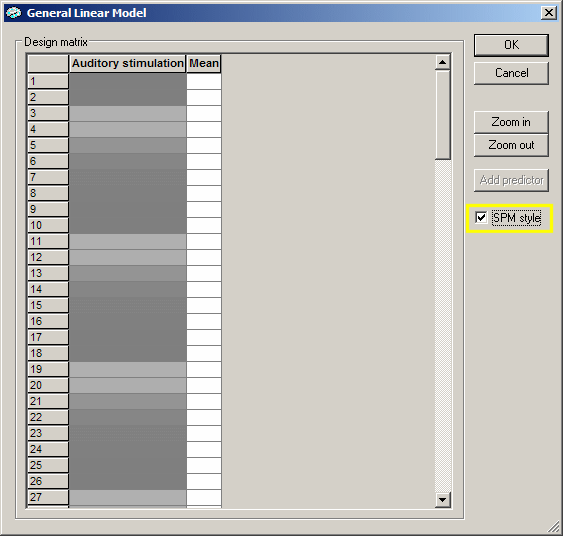
The design matrix contains only one main predictor ("Auditory stimulation") and one "confound" predictor ("Mean"). We now define two more predictors in the Single Study General Linear Model dialog. Click the Cancel button to close the design matrix dialog and again to close the Options dialog. In the Single Study General Linear Model dialog, click the Add pred button which inserts a second predictor. Click with the right mouse button one of the green segments or on "Clock imagery" in the Stimulation Protocol dialog. Now click the HRF button to apply the hemodynamic response function to the new predictor. Repeat the described steps again to define also the third predictor "Response" (blue segments). You should now have a display similar like the following one:

The Predictors field reflects now that three predictors have been defined ("/3") and that the third predictor is currently displayed. You can browse through the definitions of all three predictors by using the predictor spin box (see yellow rectangle in the figure above). We now have completed the design matrix. To visualize it numerically, we invoke again the design matrix dialog by clicking the Options button and then the Design matrix button. After checking the SPM style option, the design matrix will look as shown below:
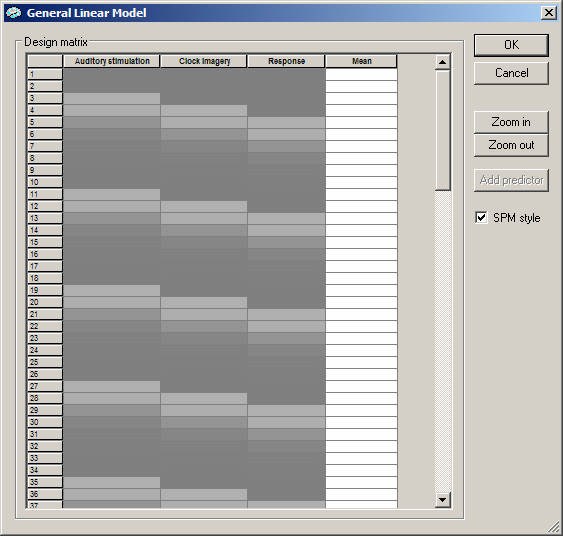
We see now the three main predictors in the first three columns and the confound ("Mean") predictor in the fourth column. Close the dialog as well as the Options dialog by clicking on the respective Cancel buttons.
Design matrix file format. Since a single study design matrix might be used later, i.e. when constructing a multi study design matrix, it is useful to save the design matrix to disk. Click the Save reference time course... item in the local File menu of the Single Study General Linear Model dialog. In the appearing Save As dialog, enter "ClockTaskDesignMatrix_Run1" as the file name and then click the Save button. To see how the design matrix has been stored in the file, open it in any ASCII editor, for example Notepad:

Following a small header with information
about the file version etc., the design matrix is displayed in columns
in the same way as in the design matrix dialog. The file contains only
the three main predictors since any level ("Mean") confound
predictors are added by the program automatically prior to any GLM computation.
The design matrix is stored in an RTC file which is the file type in BrainVoyager
to save time course data of regions-of-interest as well as for statistical
models. To clearly separate whether a time course is saved from a ROI
or from a statistical model, the RTC file format has been improved since
BrainVoyager v4.4 to reflect this in the header. The header entry "Type:"
is now used to specify the source of the subsequent time series, in this
case "DesignMatrix". Based on the type of the RTC file, subsequent
informations are parsed accordingly. The new RTC file format, thus allows
it now to save also the names of the predictors as shown in the above
text file.
Note RTC
files created with a program version prior to 4.4 can still be used in
the new version. Since they can not represent predictor file names, we
recommend, however, to switch to the new file type. This can be done simply
by loading an old RTC file or by redefining the predictors, naming them
appropriately and saving the design matrix to disk.
Note You
can use the described file format to create design matrix files outside
of BrainVoyager, for example in the case you want to use your own hemodynamic
response function. If you save the resulting design matrix in the simple
file format shown above, you can easily load it into the Single
Study General Linear Model dialog by using the Load
reference time course... entry in its local File
menu. Note that the predictor names have to be put in (") signs to
allow for names containing blanks.
Multi study design matrices
Multi study design matrices are constructed from a set of single study
design matrices. Therefore, you must first specify the single-study design
matrices of all studies which you want to include in the multi study analysis.
A multi study GLM analysis is defined by simply referring to time course
data files (FMRs or VTCs) and associated single study design matrix files
(RTCs). From this information, a proper multi study design matrix file
is automatically constructed as described below. BrainVoyager offers three
ways to construct a multi study design matrix from a series of single-study
design matrices:
One set of predictors across all studies (concatenation of predictors)
Creation of a separate set of predictors for each subject
Creation of a separate set of predictors for each study
The necessary specifications are performed in the General Linear Model: Multi Study, Multi Subject dialog as described below.
To explain the details of the creation of multi study design matrices, we will use the "mental clock task" example as described above (section "Single-study design matrices"). Close any open projects. Open the "BS_TAL.vmr" project file located in the "ClockTask" folder and then select the General Linear Model: Multi study... item in the Analysis menu. This will invoke the General Linear Model: Multi Study, Multi Subject dialog:

The large central region of the dialog shows the multi study list box which is empty at present. When filled, each row of this list box will refer to a single study (FMR or VTC file name) together with an associated single-study design matrix (RTC file name). The multi study list box is filled sequentially with pairs of single-study data and design matrix files. In this example, we will include four studies, two runs from two subjects. Click the Add... button to specify the necessary files for the first study. The appearing Open dialog asks for a VTC file. Select the file "BS_run1_r1_pp.vtc" which contains the data of the first run of the first subject and click the Open button. A second Open dialog appears asking for a single study design matrix (RTC) file. Select the file "ClockTaskDesignMatrix_run1.rtc" and click the Open button. The first row of the multi study list box will now be filled as shown below:

The first column of the first row refers to one VTC data file to be included in the multi study GLM. The second column of the first row contains the single study design matrix file (.RTC) specifying the statistical model to be used for the data file in the first column. The last column shows the names of the predictors found in the RTC file. The predictor names are shown so that you can check that each subsequently included study uses the same predictors in the same order which is a prerequisite for any multi study GLM.
Remark Each included study must use the same predictors defined in the same order so that the program can combine predictors with the same "meaning" (i.e. same condition) across studies. The defined time course of corresponding single study predictors (condition "A" in study 1, condition "A" in study 2 etc) can be, however, different for each study; this allows, for example, to present individual trials in randomized order and/or to balance the order of trials across runs and/or subjects. In the example experiment, the time points and the order of the trials in the different runs were identical, thus, we could use the same single study design matrix file for all runs/subjects. More generally, however, you might want to use a different design matrix file for each single run.
As with single studies, we can check at any time the current state of the internally built multi study design matrix. Simply click the Design matrix button in the right lower part of the General Linear Model: Multi Study, Multi Subject dialog to invoke the design matrix dialog.

The design matrix looks exactly identical
to the one we have created interactively in the Single
Study General Linear Model dialog (see above). This is, of course,
to be expected since we have included just one study so far and specified
the design matrix file we have saved previously. If we would run the multi
study GLM right now, it would, thus, compute exactly the same result as
the single study GLM.
Click the Cancel button to close
the design matrix dialog. We now add three more studies by clicking three
times the Add... button, each
time specifying another "data file - design matrix file" pair.
Add the VTC file for run 2 of subject "BS" with the respective
design matrix file. Then add the files for run 1 and 2 of subject "ML".
There also the files for run 3 and 4 in the "ClockTask" folder
but we will not use these files here. The dialog should now look as shown
in the figure below:

Let's now inspect the internally built multi study design matrix by clicking the Design matrix button. After clicking the Zoom out button several times, the design matrix should look as shown in the following figure:

The three main predictors are shown in
the first three columns as before, however they extend now over all four
runs. In other words, the multi study design matrix has been created by
concatenating the four single
study design matrices. We have, thus, one set of predictors across all
studies. This is one of three ways how the program constructs a multi
study design matrix, the other two ways using separate sets of predictors
for each study or subject will be described shortly. In the figure above
the design matrix segments for the first two studies have been labelled
with yellow brackets ("Study 1", "Study 2"), the time
points of the third study are only partially visible and the fourth study
is not visible in the figure. You can, however, browse with the vertical
scroll bar to any section of the design matrix.
Note, that BrainVoyager has added four signal level "confound"
predictors automatically. Each of these predictors is set to a value of
1.0 (white color) for all measurement time points of a study and to 0.0
(grey color) for all other studies.
Remark The different constant terms for the different studies allow each study to have a different signal level. This is very important since different runs of different subjects - and even of the same subject in the same scanning session - normally produce different signal levels. A visualization of these study level effects can be obtained for any region-of-interest with the ROI GLM analysis tool. If you check the "z-transform" option in the General Linear Model: Multi Study, Multi Subject dialog, the signal level confounds will be estimated as 0.0 because the mean of the signal time courses will become zero after a z transformation. In this case, the study signal level confounds could be removed from the design matrix [but they remain included in the present version of BrainVoyager]. If you uncheck the "z-transform" option, each study constant predictor will be estimated as the mean of the data points belonging to the respective study.
The concatenation of the predictors means that the signal changes at a voxel time course are estimated by the same three values (beta weights of the three main predictors) across the concatenated data points. This is also reflected in the resulting GLM where you will find three main predictors for specifying contrasts. To check this, close the design matrix dialog and run the GLM by clicking the GO button. If you then invoke the Overlay GLM Contrasts and Contribution Maps dialog, it will look as follows:

The three filled rows represent the three
main predictors of the multi study design matrix. You can now specify
contrasts and compute statistical maps showing voxels where a contrast
reaches significance across the
four studies.
The concatenation approach to built a multi study design matrix is a reasonable
approach if the included studies are multiple runs of the same subject.
If runs from multiple subjects are used, the results will not reach highly
significant levels at non-aligned brain regions. Using spatial smoothing,
the likelihood to have corresponding active regions across subjects can
be increased [but this approach has problems which will not discussed
here]. To obtain better insights how multiple studies contribute to an
overall statistical map, it is possible to estimate a separate
set of predictors for each included study. After running the GLM, this
allows to compute statistical maps for each individual study as well as
for any set of combined studies. In addition, this approach allows to
specify study x predictor interaction effects. The only change we have
to do to switch from the concatenation approach to the separate study
predictors approach is to check the Separate
study predictors option in the General
Linear Model: Multi Study, Multi Subject dialog:

We can now inspect the multi study design matrix reflecting the separate study predictor settings. Click the Design matrix button. After clicking the Zoom out button several times, the design matrix should look as shown in the following figure:

As you can see, there are now four sets of the three main predictors. Each predictor set defines a time course (non-zero values) only for one study but contains zero values (grey color) for all other studies. Therefore it can be said that each study has its own set of separated predictors. As before, the design matrix segments for the first two studies have been labelled with yellow brackets ("Study 1", "Study 2"), the time points of the third study are only partially visible and the fourth study is not visible in the figure.
The separation of predictors for each study means that the signal changes at a voxel time course are estimated by 12 (4*3) values (beta weights of the main predictors) across the concatenated data points plus the respective constant terms. Since the predictors for a particular study contain, however, only zero values for the other studies, only four values (beta weights of three main predictors plus constant term) actually estimate the time course of a single study. This is also reflected in the resulting GLM where you will find 12 main predictors for specifying contrasts. To check this, close the design matrix dialog and run the GLM by clicking the GO button. If you then invoke the Overlay GLM Contrasts and Contribution Maps dialog, it will look as follows:

The twelve filled rows represent the three main predictors for each of the four studies of the multi study design matrix. The three main predictors are appropriately labelled to reflect the study to which they belong. You can now specify contrasts within any single study or across any set of studies providing more flexibility than was available with the concatenation approach. The signal level confound predictors (constant term for each study) are not shown as default. If you want to see these four predictors, click the Options button and check the Show option in the Confound predictors field.
The third way to build a multi study design matrix from a set of single-study design matrices is the separate subject predictors approach. This approach is very similar to the separate study predictors approach but it pools the predictors of all studies (runs) belonging to a subject. In the case that only one run was performed for each subject, the separate subject predictors approach is identical to the separate study predictors approach. The separate subject predictors approach is required for a random effects analysis. In order to be able to group the studies to subjects accordingly, BrainVoyager uses a specific naming scheme which is described at the beginning of the section "Random effects analysis". The only change we have to do to switch to the separate subject predictors approach is to check the Separate subject predictors option in the General Linear Model: Multi Study, Multi Subject dialog:

We can now inspect the multi study design matrix reflecting the separate subject predictor settings. Click the Design matrix button. After clicking the Zoom out button several times, the design matrix should look as shown in the following figure:

As you can see, there are now two sets of the three main predictors. Each predictor set defines a time course (non-zero values) for two studies each belonging to one subject but contains zero values (grey color) for all other studies. Each subject, thus, has its own set of separated predictors. As before, the design matrix segments for the first two studies have been labelled with yellow brackets ("Study 1", "Study 2"), the time points of the third study are only partially visible and the fourth study is not visible in the figure.
The separation of predictors for each subject means that the signal changes at a voxel time course are estimated by 6 (2*3) values (beta weights of the main predictors) across the concatenated data points plus the respective constant terms. Since the predictors for a particular study contain, however, only zero values for the other studies, only four values (beta weights of the three main predictors plus constant term) actually estimate the time course of the studies belonging to a single subject. Although the runs belonging to the same subject are explained by one set of predictors, the signal level confounds are still defined separately for each study (see figure below). This is also reflected in the resulting GLM where you will find 6 main predictors for specifying contrasts and 4 confound predictors. To check this, close the design matrix dialog and run the GLM by clicking the GO button. If you then invoke the Overlay GLM Contrasts and Contribution Maps dialog, it will look as follows:

The signal level confound predictors are normally hidden but have been enabled by checking the Show option in the Confound predictors field of the Options dialog. The first six rows represent the three main predictors for each subject of the multi study design matrix. The three main predictors are appropriately labelled to reflect the subject to which they belong. You can now specify contrasts within any single subject or across subjects providing more flexibility than was available with the concatenation approach.
Note To simplify the specification of the same contrast for each subject, hold down the CTRL key while specifying with left and right mouse button clicks the respective contrast for one subject. The pressed CTRL key ensures that the defined contrast is copied to all other subjects.
Multi study design matrix definition files. After having interactively specified the data files and single-study design matrix files to be included in the multi study analysis, you might want to save the multi study specification to disk for later use. To save or load multi study specifications, you can use the Save multi study definition file... and Load multi study definition file... entries in the local File menu of the General Linear Model: Multi Study, Multi Subject dialog. A saved multi study design matrix (MDM) file contains a list of pairs with the names of the VTCs (or FMRs) and single-study design matrix files as in the multi study GLM dialog. The only difference is that full path names are written to the file and that there is a small header containing flags to specify whether z-transformation of the voxel time courses should be used in a subsequent GLM computation and which approach to built the multi study design matrix (0 = concatenation approach, 1 = separate study predictor approach, 2 = separate subject predictor approach). After loading a MDM file, these settings can be changed, of course, in the multi study GLM dialog. A MDM file for the used example in this section looks like the following:
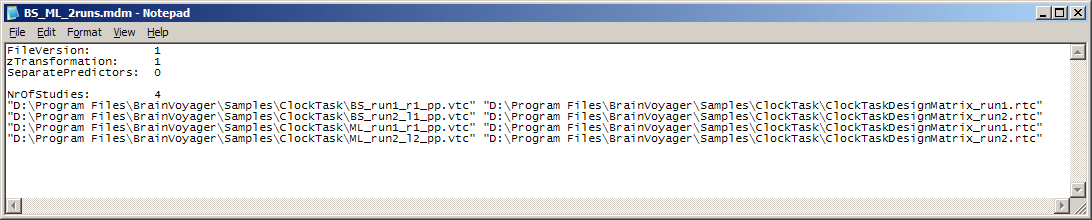
Note Besides saving an interactively specified multi study analysis, advanced users might also want to use own programs to create MDM files outside of BrainVoyager (i.e. for scripting purposes).
Note MDM files must be saved also to be able to use the multi study ROI GLM analysis tool.